Genetic genealogy is the application of genetics to traditional genealogy and involves the use of genealogical DNA testing to determine the level of genetic relationship between individuals. A genealogical DNA test examines the nucleotides at specific locations on a person's DNA for genetic genealogy purposes. The test results are not meant to have any informative medical value and do not determine specific genetic diseases or disorders; they are intended only to give genealogical information. Genealogical DNA tests generally involve comparing the results of living individuals to historic populations.
Deoxyribonucleic acid
Deoxyribonucleic acid (DNA) is a nucleic acid (i.e. a macromolecule comprising chains of monomeric nucleotides) that contains the genetic instructions used in the development and functioning of all known living organisms and some viruses. The main rôle of DNA molecules is the long-term storage of information. DNA is often compared to a set of blueprints or a recipe, or a code, since it contains the instructions needed to construct other components of cells, such as proteins and RNA molecules. The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in regulating the use of this genetic information.
Chemically, DNA consists of two long polymers of simple units called nucleotides, with backbones made of sugars and phosphate groups joined by ester bonds. Nucleotides are molecules that, when joined together, make up the structural units of RNA and DNA. In addition, nucleotides play central roles in metabolism. In that capacity, they serve as sources of chemical energy, participate in cellular signalling and are incorporated into important cofactors of enzymatic reactions. These two DNA polymer strands run in opposite directions to each other and are therefore anti-parallel. Attached to each sugar is one of four types of molecules called bases. It is the sequence of these four bases along the backbone that encodes information. This information is read using the genetic code, which specifies the sequence of the amino acids within proteins. The code is read by copying stretches of DNA into the related nucleic acid RNA, in a process called transcription.
Within cells, DNA is organised into long structures called chromosomes. These chromosomes are duplicated before cells divide, in a process called DNA replication. Eukaryotic organisms (animals, plants, fungi, and protists) store most of their DNA inside the cell nucleus and some of their DNA in organelles, such as mitochondria or chloroplasts. In contrast, prokaryotes (bacteria and archaea) store their DNA only in the cytoplasm. Within the chromosomes, chromatin proteins such as histones compact and organise DNA. These compact structures guide the interactions between DNA and other proteins, helping control which parts of the DNA are transcribed.
Paternal and maternal lineages via DNA testing
The two most common types of genetic genealogy tests are Y-DNA (paternal line) and mtDNA (maternal line) genealogical DNA tests. Note that the terms "Y chromosome" and "Y-DNA" are used interchangeably on this webpage.
The Y chromosome is one of the two sex-determining chromosomes in most mammals, including humans. In mammals, it contains the gene SRY, which triggers testis development if present. The human Y chromosome is composed of about 60 million base pairs. Females do not have a Y chromosome. DNA in the Y chromosome is passed from father to son, thus tracking many surnames. Y-DNA analysis is therefore used in family history research.
Mitochondrial DNA (mtDNA) is the DNA located in organelles called mitochondria, structures within eukaryotic cells that convert the energy from food into a form that cells can use. Most other DNA present in eukaryotic organisms is found in the cell nucleus.
Nuclear and mitochondrial DNA are thought to be of separate evolutionary origin, with the mtDNA being derived from the circular genomes of the bacteria that were engulfed by the early ancestors of today's eukaryotic cells. Each mitochondrion is estimated to contain 2-10 mtDNA copies. In the cells of extant organisms, the vast majority of the proteins present in the mitochondria (numbering approximately 1,500 different types in mammals) are coded for by nuclear DNA, but the genes for some of them, if not most, are thought to have originally been of bacterial origin, having since been transferred to the eukaryotic nucleus during evolution.
In most multicellular organisms, mtDNA is inherited from the mother (maternally inherited). Mechanisms for this include:
- simple dilution (an egg contains 100,000 to 1,000,000 mtDNA molecules, whereas a sperm contains only 100 to 1,000)
- degradation of sperm mtDNA in the fertilised egg,
- and, at least in a few organisms, failure of sperm mtDNA to enter the egg.
Whatever the mechanism, this single parent (uniparental) pattern of mtDNA inheritance is found in most animals, most plants and in fungi as well. mtDNA is particularly susceptible to reactive oxygen species generated by the respiratory chain due to its close proximity. Though mtDNA is packaged by proteins and harbours significant DNA repair capacity, these protective functions are less robust than those operating on nuclear DNA and therefore thought to contribute to enhanced susceptibility of mtDNA to oxidative damage. Mutations in mtDNA can in some cases cause maternally inherited diseases and some evidence suggests that they might be major contributors to the aging process and age-associated pathologies.
In humans (and probably in metazoans in general), 100-10,000 separate copies of mtDNA are usually present per cell (egg and sperm cells are exceptions). In mammals, each double-stranded circular mtDNA molecule consists of 15,000-17,000 base pairs. The two strands of mtDNA are differentiated by their nucleotide content with the guanine rich strand referred to as the heavy strand, and the cytosine rich strand referred to as the light strand. The heavy strand encodes 28 genes, and the light strand encodes 9 genes for a total of 37 genes. Of the 37 genes, 13 are for proteins (polypeptides), 22 are for transfer RNA (tRNA) and two are for the small and large subunits of ribosomal RNA (rRNA). This pattern is also seen among most metazoans, although in some cases one or more of the 37 genes is absent and the mtDNA size range is greater. Even greater variation in mtDNA gene content and size exists among fungi and plants, although there appears to be a core subset of genes that are present in all eukaryotes (except for the few that have no mitochondria at all). Some plant species have enormous mtDNAs (as many as 2,500,000 base pairs per mtDNA molecule) but, surprisingly, even those huge mtDNAs contain the same number and kinds of genes as related plants with much smaller mtDNAs.
Female mitochondrial inheritance
In sexual reproduction, mitochondria are normally inherited exclusively from the mother. The mitochondria in mammalian sperm are usually destroyed by the egg cell after fertilisation. Also, most mitochondria are present at the base of the sperm's tail, which is used for propelling the sperm cells. Sometimes the tail is lost during fertilisation. In 1999 it was reported that paternal sperm mitochondria (containing mtDNA) are marked with ubiquitin to select them for later destruction inside the embryo. Some in vitro fertilisation techniques, particularly injecting a sperm into an oocyte, may interfere with this.
The fact that mitochondrial DNA is maternally inherited enables researchers to trace maternal lineage far back in time. (Y-DNA, paternally inherited, is used in an analogous way to trace the agnate lineage.) This is accomplished in humans by sequencing one or more of the hypervariable control regions (HVR1 or HVR2) of the mitochondrial DNA, as with a genealogical DNA test. HVR1 consists of about 440 base pairs. These 440 base pairs are then compared to the control regions of other individuals (either specific people or subjects in a database) to determine maternal lineage. Most often, the comparison is made to the revised Cambridge Reference Sequence. Researchers have traced the matrilineal descent of domestic dogs to wolves. The concept of the Mitochondrial Eve is based on the same type of analysis, attempting to discover the origin of humanity by tracking the lineage back in time.
Because mtDNA is not highly conserved and has a rapid mutation rate, it is useful for studying the evolutionary relationships (phylogeny) of organisms. Biologists can determine and then compare mtDNA sequences among different species and use the comparisons to build an evolutionary tree for the species examined.
Because mtDNA is transmitted from mother to child (both male and female), it can be a useful tool in genealogical research into a person's maternal line.
Maternal ancestry
A person's maternal ancestry is traced by mitochondrial DNA or mtDNA for short. Both men and women possess mtDNA, but only women pass it on to their children.
So we all inherit our mtDNA from our mothers, but not from our fathers. Your mother inherited it from her mother, who inherited it from hers, and so on back through time. Therefore, mtDNA traces an unbroken maternal line back through time for generation upon generation far further back than any written record.
Research over many years has shown that all of our maternal lines are connected at some time in the past and that these connections can be traced by reading mtDNA. One striking finding was that people tended to cluster into a small number of groups, which could be defined by the precise sequence of their mtDNA. In native Europeans, for example, there were seven such groups, among Native Americans there were four, among Japanese people there were nine, and so on. Each of these groups, by an astounding yet inescapable logic, traced back to just one woman, the common maternal ancestor of everyone in her group, or clan.
DNA changes very slowly over time and this is what is used to calculate how long ago the clan mothers lived. By studying features of the geographical distribution of their present-day descendants, it can be worked out where they lived as well.
Everyone in the same clan is a direct maternal descendant of one of these clan mothers and carries her DNA within every cell of their body. Your mtDNA actually helps cells use oxygen so you are using your clan mother's mtDNA every time you breathe. However, not everyone in the same clan has exactly the same mtDNA, because DNA changes gradually over the generations. From your precise DNA test result, your place within the genealogy of the clan can be assigned.
The clan mothers were not the only people alive at the time, of course, but they were the only ones to have direct maternal descendants living right through to the present day. The other women around, or their descendants, either had no children at all or had only sons, who could not pass on their mtDNA. And, of course, the clan mothers had ancestors themselves. Amazingly, their genealogies have also been discovered. They show how everyone alive on the planet today can trace their maternal ancestry back to just one woman. By all accounts, she lived in Africa about 150,000 to 200,000 years ago and is known as "Mitochondrial Eve".
The European Clans - The seven daughters of Eve
The clan of Ursula (Latin for she-bear) is the oldest of the seven native European clans. It was founded around 45,000 years ago by the first modern humans, Homo sapiens, as they established themselves in Europe. Today, about 11% of modern Europeans are the direct maternal descendants of Ursula. They come from all parts of Europe, but the clan is particularly well represented in western Britain and Scandinavia.
The clan of Xenia (Greek for hospitable) is the second oldest of the seven native European clans. It was founded 25,000 years ago by the second wave of modern humans, Homo sapiens, who established themselves in Europe, just prior to the coldest part of the last Ice Age. Today around 7% of native Europeans are in the clan of Xenia. Within the clan, three distinct branches fan out over Europe. One is still largely confined to Eastern Europe while the other two have spread further to the West into central Europe and as far as France and Britain. About 1% of Native Americans are also in the clan of Xenia.
The clan of Helena (Greek for light) is by far the largest and most successful of the seven native clans with 41% of Europeans belonging to one of its many branches. It began 20,000 years ago with the birth of Helena somewhere in the valleys of the Dordogne and the Vezere, in south-central France. The clan is widespread throughout all parts of Europe, but reaches its highest frequency among the Basque people of northern Spain and southern France.
The clan of Velda (Scandinavian for ruler) is the smallest of the seven clans containing only about 4% of native Europeans. Velda lived 17,000 years ago in the limestone hills of Cantabria in northwest Spain. Her descendants are found nowadays mainly in western and northern Europe and are surprisingly frequent among the Saami people of Finland and Northern Norway.
The clan of Tara (Gaelic for rocky hill) includes slightly fewer than 10% of modern Europeans. Its many branches are widely distributed throughout southern and western Europe with particularly high concentrations in Ireland and the west of Britain. Tara herself lived 17,000 years ago in the northwest of Italy among the hills of Tuscany and along the estuary of the river Arno.
The clan of Katrine (Greek for pure) is a medium sized clan with 10% of Europeans among its membership. Katrine herself lived 15,000 years ago in the wooded plains of northeast Italy, now flooded by the Adriatic, and among the southern foothills of the Alps. Her descendants are still there in numbers, but have also spread throughout central and northern Europe.
The clan of Jasmine (Persian for flower) is the second largest of the seven European clans after Helena and is the only one to have its origins outside Europe. Jasmine and her descendants, who now make up 12% of Europeans, were among the first farmers and brought the agricultural revolution to Europe from the Middle East around 8,500 years ago.
The clan of Ulrike (German for Mistress of All) is not among the original "Seven Daughters of Eve" clans, but with just under 2% of Europeans among its members, it has a claim to being included among the numerically important clans. Ulrike lived about 18,000 years ago in the cold refuges of the Ukraine at the northern limits of human habitation. Though Ulrike's descendants are nowhere common, the clan is found today mainly in the east and north of Europe with particularly high concentrations in Scandinavia and the Baltic states.
Male mitochondrial inheritance
It has been reported that mitochondria can occasionally be inherited from the father in some species such as mussels. Paternally inherited mitochondria have additionally been reported in some insects such as fruit flies, honeybees, and periodical cicadas.
Evidence supports rare instances of male mitochondrial inheritance in some mammals as well. Specifically, documented occurrences exist for mice, where the male-inherited mitochondria was subsequently rejected. It has also been found in sheep, and in cloned cattle. It has been found in a single case in a human male and was linked to infertility.
While many of these cases involve cloned embryos or subsequent rejection of the paternal mitochondria, others document in vivo inheritance and persistence under lab conditions.
Paternal ancestry
A person's paternal ancestry can be traced by DNA on the Y-Chromosome (Y-DNA). Only men have a Y-Chromosome, which they inherited from their fathers and will pass on to their sons.
However, women can easily find out about their paternal ancestry from the Y-DNA of a male relative, their father or brother, for instance. The Y-DNA traces a man's unbroken paternal line back way into the past in the same manner that your maternal ancestry is traced by mtDNA .
Scientific research throughout the world has shown that all our paternal lines are connected somewhere in the past and that these connections can be traced by reading the Y-DNA. As with maternal genealogies defined by mtDNA, men tend to cluster into a small number of groups, 18 in total, which can be defined by the genetic fingerprints of their Y-DNA. In native Europeans, for example, there are 5 such groups, among Native Americans there are 4, among Japanese people there are 5, and so on. The men within each of these groups are all ultimately descended from just one man, their clan father. Obviously, these ancestral clan fathers were not the only men around at the time, but they were the only ones to have direct male descendants living today. The other men around, or their descendants, had either no children at all or only daughters. These clan fathers also had male ancestral lines and these ultimately converge on the common paternal ancestor of every man alive today. This man, know as "Y-Chromosome Adam", lived in Africa 60,000 to 80,000 years ago.
Use in identification
In humans, mitochondrial DNA spans 16,569 DNA building blocks (base pairs), representing a fraction of the total DNA in cells. Unlike nuclear DNA, which is inherited from both parents and in which genes are rearranged in the process of recombination, there is usually no change in mtDNA from parent to offspring. Although mtDNA also recombines, it does so with copies of itself within the same mitochondrion. Because of this and because the mutation rate of animal mtDNA is higher than that of nuclear DNA, mtDNA is a powerful tool for tracking ancestry through females (matrilineage) and has been used in this role to track the ancestry of many species back hundreds of generations.
Human mtDNA can also be used to help identify individuals. Forensic laboratories occasionally use mtDNA comparison to identify human remains, and especially to identify older unidentified skeletal remains. Although, unlike nuclear DNA, mtDNA is not specific to one individual, it can be used in combination with other evidence (anthropological evidence, circumstantial evidence, and the like) to establish identification. mtDNA is also used to exclude possible matches between missing persons and unidentified remains. Many researchers believe that mtDNA is better suited to identification of older skeletal remains than nuclear DNA because the greater number of copies of mtDNA per cell increases the chance of obtaining a useful sample, and because a match with a living relative is possible even if numerous maternal generations separate the two:
- American outlaw Jesse James's remains were identified using a comparison between mtDNA extracted from his remains and the mtDNA of the son of the female-line great-granddaughter of his sister;
- the remains of Alexandra Feodorovna (Alix of Hesse), last Empress of Russia, and her children were identified by comparison of their mtDNA with that of Prince Philip, Duke of Edinburgh, whose maternal grandmother was Alexandra's sister Victoria of Hesse;
- to identify Emperor Nicholas II remains his mitochondrial DNA was compared with that of James Carnegie, 3rd Duke of Fife, whose maternal great-grandmother Alexandra of Denmark (Queen Alexandra) was sister of Nicholas II mother Dagmar of Denmark (Empress Maria Feodorovna).
The low effective population size and rapid mutation rate (in animals) makes mtDNA useful for assessing genetic relationships of individuals or groups within a species and also for identifying and quantifying the phylogeny (evolutionary relationships - phylogenetics) among different species, provided they are not too distantly related. To do this, biologists determine and then compare the mtDNA sequences from different individuals or species. Data from the comparisons is used to construct a network of relationships among the sequences, which provides an estimate of the relationships among the individuals or species from which the mtDNAs were taken. This approach has limits that are imposed by the rate of mtDNA sequence change. In animals, the rapid rate of change makes mtDNA most useful for comparisons of individuals within species and for comparisons of species that are closely or moderately-closely related, among which the number of sequence differences can be easily counted. As the species become more distantly related, the number of sequence differences becomes very large; changes begin to accumulate on changes until an accurate count becomes impossible.
These tests involve the comparison of certain sequences of the DNA of pairs of individuals in order to estimate the probability that they share a common ancestor in a genealogical time frame and, through the use of a Bayesian model published by Bruce Walsh, to estimate the number of generations separating the two individuals from their most recent common ancestor or "MRCA".
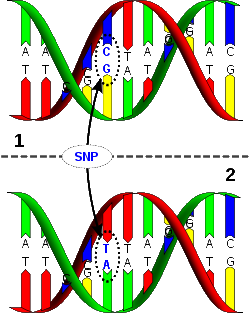 |
Y-DNA testing involves short tandem repeat (STR) and, sometimes, single nucleotide polymorphism (SNP) testing of the Y-chromosome. The STR segments which are examined are referred to as genetic markers and occur in what is considered "junk" DNA. The number of repetitions varies from one person to another and a particular number of repetitions is known as an allele of the marker. An STR on the Y chromosome is designated by a DYS number (Y-DNA Segment number). An SNP is a change to a single nucleotide in a DNA sequence. The relative mutation rate for an SNP is extremely low. This makes them ideal for marking the history of the human genetic tree. SNPs are named with a letter code and a number. The letter indicates the lab or research team that discovered the SNP. The number indicates the order in which it was discovered. For example M269 is the 269th SNP documented by the Human Population Genetics Laboratory at Stanford University, which uses the letter M. |
| Strand 1 differs from strand 2 at a single base pair location (a C-T polymorphism) |
The Y-chromosome is present only in males and reveals information on the strict paternal line. These tests can provide insight into the recent (via STRs) and ancient (via SNPs) genetic ancestry. Y-DNA tests generally examine 10-67 STR markers on the Y chromosome, but over 100 markers are available. STR test results provide the personal haplotype which should be similar among all male descendants of a male ancestor. SNP test results are used to assign people to a paternal haplogroup, which defines a much larger genetic population.
Haplotype
A Y-DNA haplotype is the numbered results of a genealogical Y-DNA test. Each allele value has a distinctive frequency within a population. For example, at DYS455, the results will show 8, 9, 10, 11 or 12 repeats, with 11 being most common. For high marker tests the allele frequencies provide a signature for a surname lineage. The test results are then compared to another project member's results to determine the time frame in which the two people shared an MRCA. If the two tests match perfectly on 37 markers, there is a 50% probability that the MRCA was fewer than 2 to 3 generations ago, 90% probability that the MRCA was fewer than 5 generations ago, and 95% probability that the MRCA was fewer than 7 generations ago. Before choosing a test, it is important for an individual to check the number of markers that will be tested. For example, the Genographic Project looks at only 12 markers, while most laboratories and surname projects recommend testing at least 25. The more markers that are tested, the more discriminating and powerful the results will be. A 12-marker STR test is usually not discriminating enough to provide conclusive results for a common surname. STRs results may also indicate a likely haplogroup, though this can only be confirmed by specifically testing for that Haplogroup's SNPs.
Haplogroup
Haplogroups are large groups of haplotypes that can be used to define genetic populations and are often geographically oriented. Y-DNA haplogroups are determined by SNP tests. SNPs are locations on the DNA where one nucleotide has "mutated" or "switched" to a different nucleotide. The nucleotide switch must occur in at least 1% of the population to be considered a useful SNP. If it occurs in less than 1% of the population, it is considered a personal SNP.
One way to think about haplogroups is as major branches on the family tree of Homo sapiens. These haplogroup branches characterise the early migrations of population groups. As a result, haplogroups are usually associated with a geographic region. If haplogroups are the branches of the tree then the haplotypes represent the leaves of the tree. All of the haplotypes that belong to a particular haplogroup are leaves on the same branch. Both mtDNA and Y-DNA tests provide haplogroup information, but the haplogroups nomenclature are different for each:
- a Y-DNA haplogroup is defined as all of the male descendants of the single person who first showed a particular SNP mutation. An SNP mutation identifies a group who share a common ancestor far back in time, since SNPs rarely mutate. Each member of a particular haplogroup has the same SNP mutation;
- an mtDNA haplogroup is defined as all of the female descendants of the single person who first showed a particular polymorphism, or SNP mutation. Like Y-DNA SNP mutations, an mtDNA SNP mutation identifies a group who share a common ancestor far back in time.
Haplogroup prediction
A person's haplogroup can often be inferred from their haplotype, but can be proven only with a Y-chromosome SNP tests (Y-SNP test). In addition, some companies offer sub-clade tests, such as for Haplogroup R. Few haplotypes will exactly match the modal values for Haplogroup R. One can consult an allele frequency table to determine the likelihood of remaining in Haplogroup R based on the variations observed. Additional predictions include:
- if DYS426 is 12 and DYS392 is 11, one is probably a member of haplogroup R1a1
- if DYS426 is 12 and DYS392 is not 11, one is probably a member of haplogroup R1b
- if DYS426 is 11, one is probably a member of haplogroup G,I, or J
- if DYS426 is 11 and DYS388 is 12, one is in the known modal haplotype for G
A Bayes classifier to predict the haplogroup probabilities for an observed haplotype is available at the Whit Athey Haplogroup Predictor website.
mtDNA testing involves sequencing or testing the HVR-1 region, HVR-2 region or both. An mtDNA test may also include the additional SNPs needed to assign people to a maternal haplogroup - or even include the complete mtDNA.
Either Y-DNA or mtDNA test results can be compared to the results of others via private or public DNA databases.
Biogeographical and ethnic origins
Additional DNA tests exist for determining biogeographical and ethnic origin, but these tests have less relevance for traditional genealogy.
Genetic genealogy has revealed astonishing links between peoples. For instance, it has shown that the ancient Phoenician people were ancestors of much of the present-day population of the island of Malta. Preliminary results from a study by Pierre Zalloua of the American University of Beirut and Spencer Wells, supported by a grant from National Geographic's Committee for Research and Exploration, were published in the October 2004 issue of National Geographic. One of the conclusions is that "more than half of the Y chromosome lineages that we see in today's Maltese population could have come in with the Phoenicians".
Human migration
Genealogical DNA testing methods are also being used on a longer time scale to trace human migratory patterns. For example, they have been used to determine when the first humans came to North America and what path they followed.
For several years, a number of researchers and laboratories from around the world have been sampling indigenous populations from around the globe in an effort to map historical human migration patterns. Recently, several projects have been created that are aimed at bringing this science to the public. One example is the National Geographic Society's "Genographic Project", which aims to map historical human migration patterns by collecting and analysing DNA samples from over 100,000 people across five continents. Another example is the "DNA Clans Genetic Ancestry Analysis", which measures a person's precise genetic connections to indigenous ethnic groups from around the world.
Typical customers and interest groups
Male DNA testing customers most often start with a Y chromosome test to determine their father's paternal ancestry. Females generally begin with a mitochondrial test to trace their ancient maternal lineage, which males often have tested for the same purpose.
A common consumer goal in purchasing DNA testing services is to acquire quantified, scientific linkage to a specific ancestral group. A compelling example of this motive is found in the expressed desires of some consumers to be proven to have Viking paternal ancestry. In keeping with this marketplace demand Oxford Ancestors offers a Y chromosome test purporting to assess whether given males are of "Viking stock." Those whose DNA falls into the designated haplogroup are issued Viking descendant certificates by the testing service. Oxford Associates also participated in producing the BBC televised documentary "The Blood of the Vikings" which showed how DNA testing could reveal Viking ancestry.
The RootsWeb Genealogy-DNA Internet discussion group has a membership of 750 subscribers from around the world. Some subscribers have had various DNA tests performed and are seeking advice and guidance in interpreting their results. The list also includes administrators of DNA projects that examine surnames, geographic regions, or ethnic groups. The sophistication of subscribers ranges from expert to novice. In some cases, subscribers have been credited with making useful and novel contributions to knowledge in the field of genetic genealogy.
Paternal and maternal DNA lineages
Mitochondria are small organelles that lie in the cytoplasm of eukaryotic cells, such as those of humans. Their primary purpose is to provide energy to the cell. Mitochondria are thought to be the vestigial remains of symbiotic bacteria that were once free living. One indication that mitochondria were once free living is that they contain a relatively small circular segment of DNA, called mitochondrial DNA (mtDNA). The overwhelming majority of a human's DNA is contained in chromosomes in the nucleus of the cell, but mtDNA is an exception. Individuals inherit their cytoplasm and the organelles it contains exclusively from their mothers, as these are derived from the ovum (egg cell) only, not from the sperm. When a mutation arises in mtDNA molecule, the mutation is therefore passed in a direct female line of descent. These rare mutations are derived from copying mistakes - when the DNA is copied it is possible that a single mistake occurs in the DNA sequence, an outcome which is called a single nucleotide polymorphism (SNP).
Human Y chromosomes are male-specific sex chromosomes. Nearly all humans that possess a Y chromosome will be morphologically male. Y chromosomes are therefore passed from father to son. Although Y chromosomes are situated in the cell nucleus, they only recombine with the X chromosome at the ends of the Y chromosome. The vast majority of the Y chromosome (95%) does not recombine. When mutations (SNPs and STR copying mistakes) arise in the Y chromosome they are passed down directly from father to son in a direct male line of descent. The Y-DNA and mtDNA therefore share a certain feature - they both pass down unchanged except for mutations.
The other chromosomes, autosomes and X chromosomes in women, share their genetic material (called crossing over leading to recombination) during meiosis (a special type of cell division that occurs for the purposes of sexual reproduction). Effectively this means that the genetic material from these chromosomes gets mixed up in every generation, and so any new mutations are passed down randomly from parents to offspring.
The special feature that both Y-DNA and mtDNA share, above, preserves a "written" record of their mutations because neither DNA gets mixed up or randomised - mutations remain fixed in place on both types of DNA. Furthermore, the historical sequence of these mutations can also be inferred. For example, if a set of ten Y chromosomes (derived from ten different men) contains a mutation, A, but only five of these chromosomes contain a second mutation, B, it must be the case that mutation B occurred after mutation A. Furthermore all ten men who carry the chromosome with mutation A are the direct male line descendants of the same man who was the first to carry this mutation. The first man to carry mutation B was also a direct male line descendant of this man, but is also the direct male line ancestor of all men carrying mutation B. Series of mutations such as this form molecular lineages. Furthermore each SNP mutation may define a set of specific Y chromosomes called a haplogroup. All men carrying SNP mutation A form a single haplogroup, and all men carrying mutation B are part of this haplogroup, but mutation B (if a SNP) may also define a more recent haplogroup (which is a subgroup or subclade) of its own which men carrying only mutation A do not belong to. Both mtDNA and Y chromosomes or Y-DNA are grouped into lineages and haplogroups; these are often presented as tree-like diagrams.
Benefits
Genetic genealogy gives genealogists a means to check or supplement their genealogy results with information obtained via DNA testing. A positive test match with another individual may:
- provide locations for further genealogical research
- help determine ancestral homeland
- discover living relatives
- validate existing research
- confirm or deny suspected connections between families
- prove or disprove theories regarding ancestry
Drawbacks
People who resist testing may cite one of the following concerns:
- cost
- quality of testing
- concerns over privacy issues
- loss of ethnic identity
Finally, Y-DNA and mtDNA tests each only trace a single lineage (one's father's father's father's etc. lineage or one's mother's mother's mother's etc. lineage). At 10 generations back, an individual has up to 1024 unique ancestors (fewer if ancestor cousins interbred) and a Y-DNA or mtDNA test is only studying one of those ancestors, as well as their descendants and siblings (same sexed siblings for Y-DNA or all siblings for mtDNA). However, most genealogists maintain contact with many cousins (1st, 2nd, 3rd, etc., with different surnames) whose Y-DNA and mtDNA are different, and thus can be encouraged to be tested to find additional ancestral DNA lineages.
Generation intervals and their significance
In animal breeding theory the generation interval is a crucial statistic. The shorter the generation interval, everything else being equal, the faster the rate of genetic progress per annum resulting from selection.
The generation interval for a family is defined as the average age of the parents when the children were born. This is worked out for each parent separately and the mean is then taken of these two figures. e.g. If the father's age at each of the births was 22.3, 24.4, 26.7 and 28.4 years, and the mother's age was 20.1, 22.2, 24.5 and 26.2 years, then the father's generation interval is 25.45 years, the mother's is 23.25 years and the overall average is 24.35 years. The average generation interval for a whole population at any time is the weighted average of these family figures. Family generation intervals for each family can be calculated if enough information is available. The average generation interval is usually about 30 years.
The actual generation intervals and their mean for a direct line refers only to those nominated individuals in the direct line, not the whole family. The generation interval is the age of the parent when the relevant child is born. This is obtained by subtracting the birth date of the parent from that of the child in each case. To find the average generation interval for a direct line, it is not necessary to know the date of birth of every member of the line, just the dates for the first and last members. For example, in my own family, my oldest traced ancestor, Gilbarte Butler, was born in 1573 and is 12 generations back from me in the direct line. Since I was born in 1954, the time interval between the two birth dates is: 1954 - 1573 = 381 years. If this is divided by the number of connecting links between us it gives the average generation interval for the line. i.e. 381/12 = 31.75 years.
Human generation intervals can vary widely (theoretically from about 12 to 90). Some of the causes of variation are given below:
- They are often longer in male-male-male line successions than in female-female-female ones because males remain fertile longer than females.
- Another reason for variation is differences in parity i.e. in large families the generation interval within a direct line can be affected by whether the person is the first or last born.
- The generation interval of a particular family depends on the age of the parents when the first child is born. At the present time there are a lot of unplanned teenage pregnancies on the one hand, and on the other married couples often wait until their late twenties or thirties before starting a family.
- Total family size also affects the family generation interval. The standard family in Victorian times was 12, which meant that the mother was usually in her mid-forties when the last child was born. Modern families rarely exceed 4 children.
- The family generation interval is also affected by the lengths of the spaces between each birth and whether or not they are spread evenly over the fertile period.
- Different marriage laws and customs in other countries can have a bearing on the generation intervals. e.g. child marriage and polygamy.
Haplogroup R1b
R1b (previously known as Hg1 and Eu18) is the most prolific haplogroup in Europe and its frequency changes in a cline from west (where it reaches a saturation point of almost 100% in areas of Western Ireland) to east (where it becomes uncommon in parts of Eastern Europe and virtually disappears beyond the Middle East). An R1b haplotype (a set of marker scores indicative of the haplogroup) is very difficult to interpret in that they are found at relatively high frequency in the areas where the Anglo-Saxon and Danish "invaders" originally called home (e.g. 55% in Friesland), and even up to 30% in Norway. Thus an R1b haplotype makes it very challenging to determine the origin of a family with this DNA signature.
During the Last Glacial Maximum, about 18,000 years ago, the people bearing the R1b haplogroup over wintered in Northern Spain. After the glacial retreat about 12,000 years before present, R1b began a migration to the north in large numbers, and to the east in declining numbers.
R1b probably arrived in Spain from the east 30,000 years ago among the paleolithic or "old stone age" peoples considered to be aboriginal to Europe. It is believed that everyone who is R1b is a descendant in the male line from an individual known as "the patriarch" since his descendants account for over 40% of all the chromosomes of Europe. This haplogroup is characteristic of the Basques whose language is probably that of the first R1b, and who are genetically the closest to the original R1b population (which probably amounted to only a few thousand individuals).
SNP markers U106 and U152 appear to have arisen over 5,000 years ago (probably much longer) and are found in all the descendants of the man in which each first appeared. To date it appears that U106 in Britain marks "Anglo-Saxon". Norway is about two thirds U106+; and the surrogate for the Anglo-Saxon homeland (Friesland) is about 75% U106+.
U152 is seen further east and in England only in what is known as the "Danelaw" which appears to mark Danish Viking in those who possess it. It also seems to be characteristic of the Danish Isles (although this has yet to be conclusively demonstrated) and the Danish people migrated north from Southern Sweden to Norway. Thus, some Norse will be U152+ but based on research samples only from the South-eastern part of Norway - although a larger sample may locate a more widespread distribution.
Another SNP was identified in 2008 which links all of the downstream R1b1c subclades which are derived (positive) on S116 except R1b1c9 (negative on S116). About 50% of R1b1c will be S116* meaning negative on all SNPs downstream of S116. To date individuals with this motif tend to cluster along the Atlantic facade including Denmark and Norway.
A recent mutation from DYS390=24 to 23 could explain the pattern better than a geographical attribution. A few aboriginal Shetland surnames have haplotypes that are identical to those that are overwhelmingly most frequent in certain regions of Norway (according to the YHRD database). In these instances a tentative attribution can be made that the ancestor was from Western or Northern Norway - to date the only two locations where this pattern can be observed - and which fits with the probable location of the R1b emigrants from Norway in the 800s. The only pattern so far seen to have a geographical link is DYS390=23, DYS391=11 which points to a Germanic or Scandinavian origin.
DNA analysis have been made on skeletons from Viking tombs. The mtDNA haplogroups found were the same as those found today in Europe, but with a much higher percentage of the now very rare haplogroups I and X which are each found in only 1% of the modern European population. Haplogroup I has been found in over 10% of the bodies tested from Viking cemeteries. Other studies also found mtDNA haplogroup X in Anglo-Saxon skeletons, suggesting a possible Germanic origin.
Ancient Norse appeared to belong mostly to Y-DNA haplogroups I, R1a and R1b (U106+). However, there are great disparities between the regions of Scandinavia. Denmark, along with Friesland, northern Germany and the Netherlands, have the highest incidence of haplogroup R1b. Over 40% of Swedes belong to haplogroup I1a, and another 10% to haplogroup I1c. In Norway, the three haplogroups have about the same share, but with stronger R1b concentration in the South-West and R1a in the North.
It appears that Scandinavia already shared this variety of haplogroups 2,000 years ago. The only thing that has changed over time is the increased blending between the original ethnic groups that converged in northern Europe.
Subclade R1b1b2
Subclade R1b1b2 is defined by the presence of SNP marker M269. It has been found at generally low frequencies throughout central Eurasia and with relatively high frequency among Bashkirs of the Bashkortostan and Perm region (84.0%).
This subclade is defined by the presence of the M269 marker. From 2003 to 2005 what is now R1b1b2 was designated R1b3. From 2005 to 2008 it was R1b1c.
| R-M269 | |
| Long-hand: | R1b1b2 (formerly R1b1c, R1b3) |
| Defining SNP: | M269 |
| Parent Clade: | R-P297 |
| Subclades: | R-P311 |
The members of R1b3 (or R-M269, formerly known as R1b) are believed to be the descendants of the first modern humans who entered Europe about 35,000-40,000 years ago (Aurignacian culture). Those R1b3 forebears were the people who painted the beautiful art in the caves in Spain and France. They were the modern humans who were the contemporaries - and perhaps exterminators - of the European Neanderthals.
In articles published around 2000 it was proposed that this subclade came into existence in Europe before the last Ice Age (i.e. aka Last Glacial Maximum (LGM), approximately 20,000 years ago), but more recently this scenario is no longer receiving much mainstream attention. A much newer estimate for R1b1b2 arising is around 5,000 to 8,000 years ago. It also appears increasingly to be the case that Western European R1b is dominated by R-P310, also known as R-L11. It is this Western European branch which is in turn dominated by U106 and P312, and the typical most common STR Y-DNA signature for Western Europe, the so-called Atlantic Modal Haplotype, which is also sometimes referred to as "Haplotype 15". Haplotype 15 is contrasted with "Haplotype 35", which has long been noted as a distinct type of R1b1b2, more common towards the southeast of Europe.
| 67 Y-chromosome Short Tandem Repeat (STR) markers for David R Ramsdale | ||||||||||||
| Locus | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| DYS# | 393 | 390 | 19* | 391 | 385a | 385b | 426 | 388 | 439 | 389-1 | 392 | 389-2 |
| Alleles | 13 | 23 | 14 | 11 | 11 | 14 | 12 | 13 | 11 | 13 | 13 | 28 |
| Locus | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 |
| DYS# | 458 | 459a | 459b | 455 | 454 | 447 | 437 | 448 | 449 | 464a** | 464b** | 464c** | 464d** |
| Alleles | 17 | 9 | 10 | 11 | 11 | 25 | 14 | 19 | 30 | 15 | 15 | 15 | 18 |
| Locus | 26 | 27 | 28 | 29 | 30 | 31 | 32 | 33 | 34 | 35 | 36 | 37 |
| DYS# | 460 | GATA H4 | YCA II a | YCA II b | 456 | 607 | 576 | 570 | CDY a | CDY b | 442 | 438 |
| Alleles | 11 | 11 | 19 | 23 | 16 | 15 | 18 | 17 | 36 | 38 | 12 | 12 |
| Locus | 38 | 39 | 40 | 41 | 42 | 43 | 44 | 45 | 46 | 47 |
| DYS# | 531 | 578 | 395S1a | 395S1b | 590 | 537 | 641 | 472 | 406S1 | 511 |
| Alleles | 12 | 9 | 15 | 16 | 8 | 10 | 10 | 8 | 10 | 10 |
| Locus | 48 | 49 | 50 | 51 | 52 | 53 | 54 | 55 | 56 | 57 | 58 | 59 | 60 |
| DYS# | 425 | 413a | 413b | 557 | 594 | 436 | 490 | 534 | 450 | 444 | 481 | 520 | 446 |
| Alleles | 12 | 21 | 23 | 16 | 10 | 12 | 12 | 15 | 8 | 12 | 22 | 20 | 13 |
| Locus | 61 | 62 | 63 | 64 | 65 | 66 | 67 |
| DYS# | 617 | 568 | 487 | 572 | 640 | 492 | 565 |
| Alleles | 12 | 11 | 13 | 11 | 11 | 12 | 12 |
*Also known as DYS 394
**On 5/19/2003, these values were adjusted down by 1 point because of a change in Lab nomenclature.
***A value of "0" for any marker indicates that the lab reported a null value or no result for this marker. All cases of this nature are retested multiple times by the lab to confirm their accuracy. Mutations causing null values are infrequent, but are passed on to offspring just like other mutations, so related male lineages such as a father and son would likely share any null values.
Allele: one of the different forms of a gene that can exist at a single locus. Since mutations in the allele value occur very slowly with time, one should see the same allele value for a male and his great-grandfather for example.
DYS (DNA Y-Chromosome Segment): a nomenclature system which assigns DYS numbers to newly discovered markers. They are the "names" of each marker.
Locus (plural-loci): a specific spot in the genome. A variable locus will have several possible alleles.
Test results of a deep clade test have determined my exact subclade or branch of the haplogroup to be:
R1b1a2a1a1b*: P312+ M269+ L21- L48- M153- M65- SRY2627- U106- U152- L176.2- L165- DF19- L238- Z196-
This has been analyzed for Haplogroup determination using the Single Nucleotide Polymorphism test. The analysis shows that I am positive for the following SNPs:
ZZ12_1+ YSC0000288+ YSC0000269+ YSC0000225+ YSC0000224+ YSC0000191+ YSC0000186+ YSC0000075+ V9+ V52+ V250+ V241+ V205+ V189+ V186+ V168+ S4063+ s3+ PF6524+ PF6520+ PF6509+ PF6507+ PF6506+ PF6500+ PF6498+ PF6495+ PF6494+ PF6479+ PF6477+ PF6470 + PF6469+ PF6463+ PF6438+ PF6434+ PF6432+ PF6430+ PF6425+ PF6409+ PF6272+ PF6271+ PF6270+
Haplogroup R-ZZ12_1
Haplogroup R-P312 is the descendant of the major R-P25 (aka R-M343) lineage and is the most common in Central Europe, Spain, France, Portugal, and the British Isles.
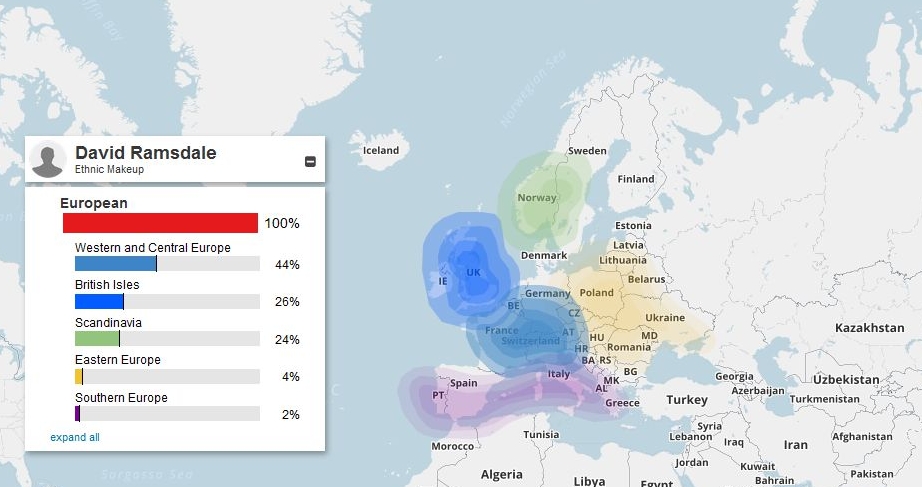
(above) DRR ethnic makeup
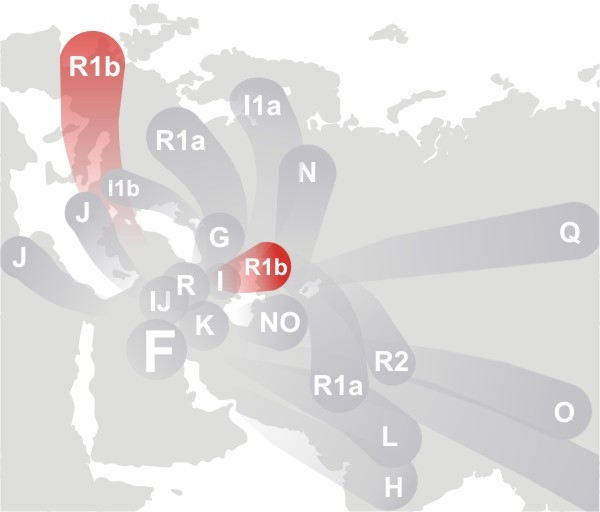
(above) DNA Migration Route Map for Haplogroup R1b
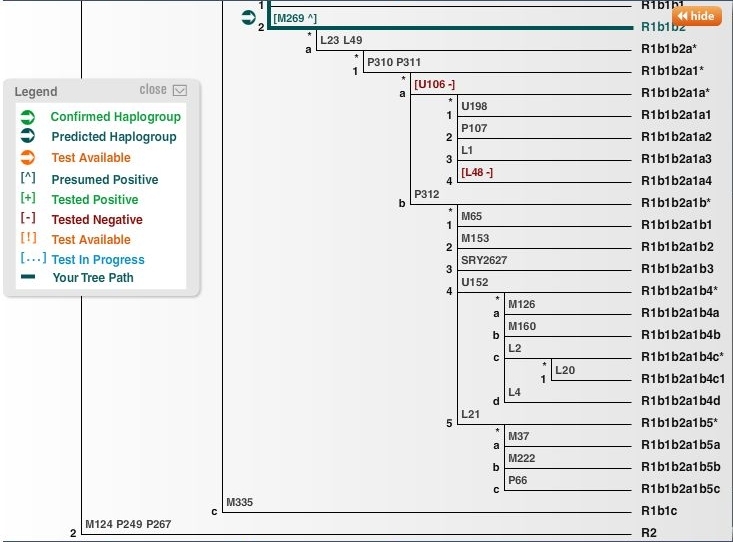
(above) DNA Y-Haplotree for Subclade R1b1b2 R-M269 (NB: tested negative for U106 & L48)
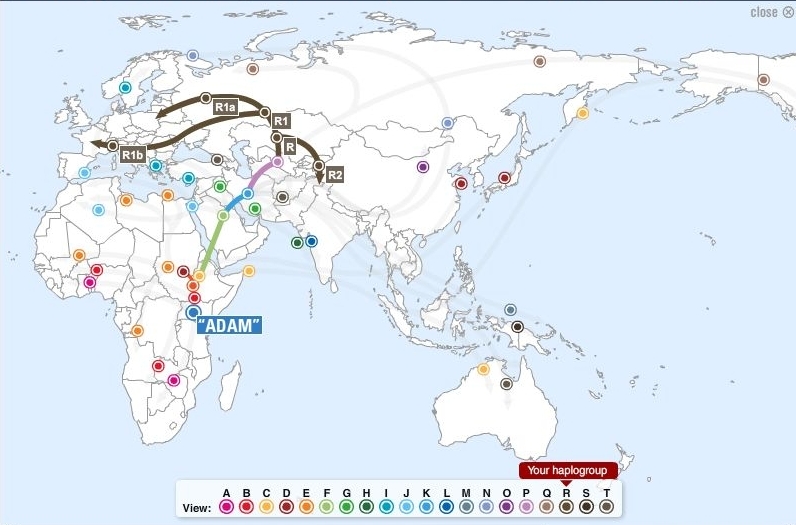
(above) DNA Migration Route Map for Haplogroup R1b
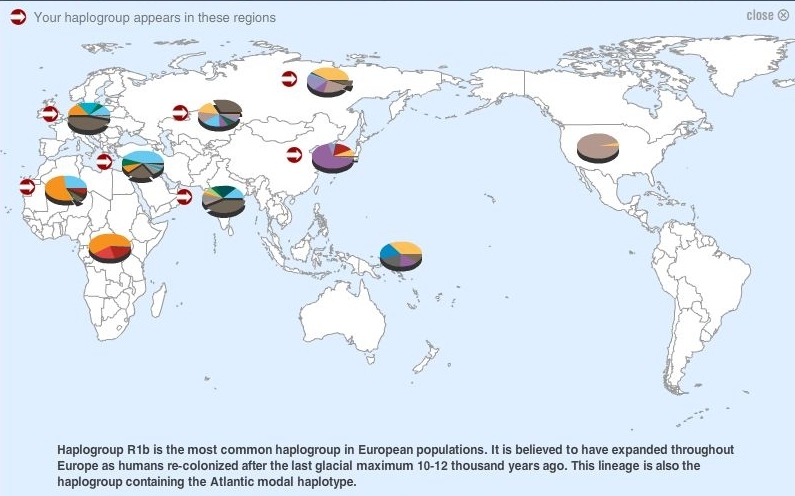
(above) DNA Frequency Map for Haplogroup R1b
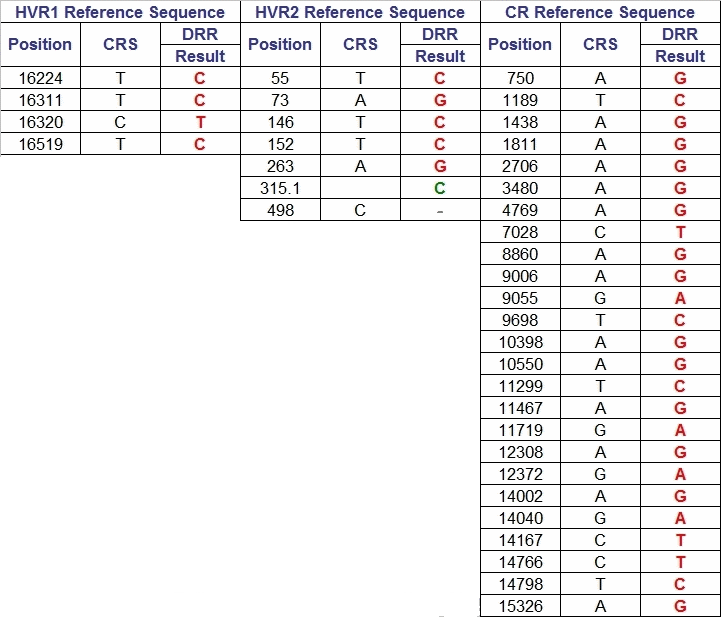
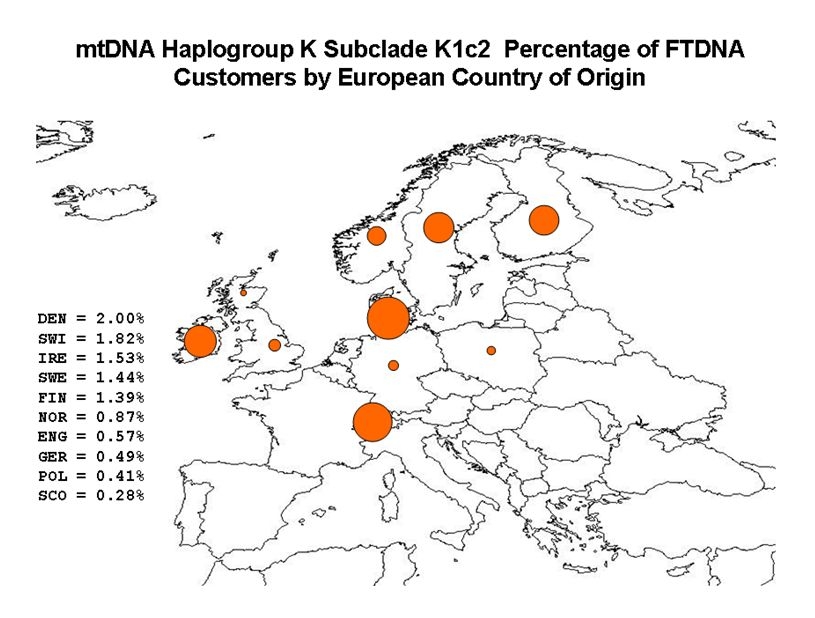
(above) My mtDNA Haplogroup (K1c2) CRS Differences
"Katrine", the founding mother of mitochondrial DNA haplogroup K, was one of the "Seven Daughters of Eve" as listed in the 2001 book of that title by Bryan Sykes. A lot has happened since 2001, but the book is still valuable. Katrine lived about 16,000 years ago. Perhaps the oldest known K descendant was Oetzi the Iceman whose frozen body was discovered in the Alps in 1991. Estimated at 5000 years old, the Iceman proved to have the basic mutations for a K: 16224C and 16311C. Every K is a cousin of Oetzi.
Although the "defining motifs" for K are unusually shown as Hyper Variable Region 1 (HVR1) mutations 16224C and 16311C, virtually all K's have 16519C in HVR1 plus 073G, 263G and 315.1C in HVR2. Those K's not shown as having those mutations likely have not been tested for them, although "back mutations" do occur. The FamilyTreeDNA mtDNAPlus test should find them all. Virtually every K will also have other mutations as part of a subclade or as personal mutations. All mutations are differences from the Cambridge Reference Sequence (CRS). There is at present no K subclade test publicly available.
Haplogroup determination using the Single Nucleotide Polymorphism test shows that David R. Ramsdale is positive for the following SNPs:
- YSC0000288+
- YSC0000269+
- YSC0000225+
- YSC0000224+
- YSC0000191+
- YSC0000186+
- YSC0000075+
- V9+
- V52+
- V250+
- V241+
- V205+
- V189+
- V186+
- V168+
- s3+
- PF6524+
- PF6509+
- PF6507+
- PF6506+
- PF6500+
- PF6498+
- PF6495+
- PF6494+
- PF6463+
- PF6438+
- PF6434+
- PF6432+
- PF6430+
- PF6425+
- PF6409+
- PF6272+
- PF6271+
- PF6270+
- PF6263+
- PF6250+
- PF6249+
- PF6246+
- PF5982+
- PF5965+
- PF5964+
- PF5957
19 February 2016
Haplogroup R-P312 is the descendant of the major R-P25 (aka R-M343) lineage and is the most common in Central Europe, Spain, France, Portugal, and the British Isles.
| 111 Y-DNA - Standard Y-STR Values for David R. Ramsdale | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PANEL 1 (1-12) | ||||||||||||
| Marker | DYS393 | DYS390 | DYS19** | DYS391 | DYS385 | DYS426 | DYS388 | DYS439 | DYS389I | DYS392 | DYS389II*** | |
| Value | 13 | 23 | 14 | 11 | 11-14 | 12 | 13 | 11 | 13 | 13 | 28 | |
| PANEL 2 (13-25) | ||||||||||||
| Marker | DYS458 | DYS459 | DYS455 | DYS454 | DYS447 | DYS437 | DYS448 | DYS449 | DYS464 | |||
| Value | 17 | 9-10 | 11 | 11 | 25 | 14 | 19 | 30 | 15-15-15-18 | |||
| PANEL 3 (26-37) | ||||||||||||
| Marker | DYS460 | Y-GATA-H4 | YCAII | DYS456 | DYS607 | DYS576 | DYS570 | CDY | DYS442 | DYS438 | ||
| Value | 11 | 11 | 19-23 | 16 | 15 | 18 | 17 | 36-38 | 12 | 12 | ||
| PANEL 4 (38-47) | ||||||||||||
| Marker | DYS531 | DYS578 | DYF395S1 | DYS590 | DYS537 | DYS641 | DYS472 | DYF406S1 | DYS511 | |||
| Value | 12 | 9 | 15-16 | 8 | 10 | 10 | 8 | 10 | 10 | |||
| PANEL 4 (48-60) | ||||||||||||
| Marker | DYS425 | DYS413 | DYS557 | DYS594 | DYS436 | DYS490 | DYS534 | DYS450 | DYS444 | DYS481 | DYS520 | DYS446 |
| Value | 12 | 21-23 | 16 | 10 | 12 | 12 | 15 | 8 | 12 | 22 | 20 | 13 |
| PANEL 4 (61-67) | ||||||||||||
| Marker | DYS617 | DYS568 | DYS487 | DYS572 | DYS640 | DYS492 | DYS565 | |||||
| Value | 12 | 11 | 13 | 11 | 11 | 12 | 12 | |||||
| PANEL 5 (68-75) | ||||||||||||
| Marker | DYS710 | DYS485 | DYS632 | DYS495 | DYS540 | DYS714 | DYS716 | DYS717 | ||||
| Value | 34 | 15 | 9 | 16 | 12 | 25 | 26 | 19 | ||||
| PANEL 5 (76-85) | ||||||||||||
| Marker | DYS505 | DYS556 | DYS549 | DYS589 | DYS522 | DYS494 | DYS533 | DYS636 | DYS575 | DYS638 | ||
| Value | 13 | 11 | 13 | 12 | 10 | 9 | 12 | 12 | 10 | 11 | ||
| PANEL 5 (86-93) | ||||||||||||
| Marker | DYS462 | DYS452 | DYS445 | Y-GATA-A10 | DYS463 | DYS441 | Y-GGAAT-1B07 | DYS525 | ||||
| Value | 11 | 30 | 12 | 13 | 24 | 13 | 10 | 10 | ||||
| PANEL 5 (94-102) | ||||||||||||
| Marker | DYS712 | DYS593 | DYS650 | DYS532 | DYS715 | DYS504 | DYS513 | DYS561 | DYS552 | |||
| Value | 20 | 15 | 18 | 14 | 24 | 17 | 12 | 15 | 24 | |||
| PANEL 5 (103-111) | ||||||||||||
| Marker | DYS726 | DYS635 | DYS587 | DYS643 | DYS497 | DYS510 | DYS434 | DYS461 | DYS435 | |||
Note: consolidation of the 111 markers results in only 102 markers being shown:
- 385 = 385a + 385b
- 459 = 459a + 459b
- 464 = 464a + 464b + 464c + 464d
- YC11 = YC11a + YC11b
- CDY = CDYa + CDYb
- DYF395S1 = 395S1a + 395S1b
- 413 = 413a + 413b